Our lab is interested in the regulation of development and antibiotic production in bacteria. We primarily study the filamentous Gram-positive actinobacterial genus Streptomyces, which have a complex life cycle that culminates in the production of chains of exospores. Streptomyces and other sporulating Actinobacteria are our most important source of antibiotics and other natural products. The production of antibiotics is tightly coordinated with the complex life cycle of these bacteria, and therefore the ability to fully exploit antibiotic production by these species depends on a profound understanding of the global regulatory cascades controlling development. Our key goals are to understand how the process of sporulation is regulated in Streptomyces, examine how these regulatory pathways have evolved in the Phylum Actinobacteria, and to leverage this knowledge to manipulate antibiotic production.

Regulation of sporulation through c-di-GMP signaling
In Streptomyces, the central integrator controlling progression through the life cycle is the intracellular nucleotide second messenger cyclic-di-GMP (c-di-GMP). c-di-GMP signaling is a universal mechanism in bacteria to control global gene expression in response to specific environmental stimuli. However, this process has almost exclusively been studied in Gram-negative bacterial models, where it controls motility and biofilm formation. Many of the components of c-di-GMP signaling networks identified in these phylogenetically distant model organisms have no known homologs in Streptomyces bacteria. We are using a number of approaches to identify and characterize proteins involved in c-di-GMP-mediated control of sporulation in Streptomyces.

Evolution of regulatory signaling networks
A remarkable diversity of lifestyles, morphologies, and metabolisms have emerged over the course of bacterial evolution. This diversification requires the concurrent modification of molecular machineries with regulatory mechanisms to maintain tight control of key cellular processes. One way this can occur is through the co-option of existing signaling networks to regulate distinct functions. c-di-GMP is an important signaling molecule across the bacteria – homologs of the proteins responsible for synthesis and degradation of c-di-GMP can be found in all major phyla. However, the function and molecular basis of c-di-GMP signaling in the majority of bacteria remains largely unknown. We use a combination of bioinformatic and molecular approaches to assess the distribution and functional diversification of c-di-GMP binding proteins across the Phylum Actinobacteria, which comprise highly diverse unicellular and filamentous species. These analyses allow us to develop hypotheses about specific functions and mechanisms of c-di-GMP signaling in diverse industrially important taxa.
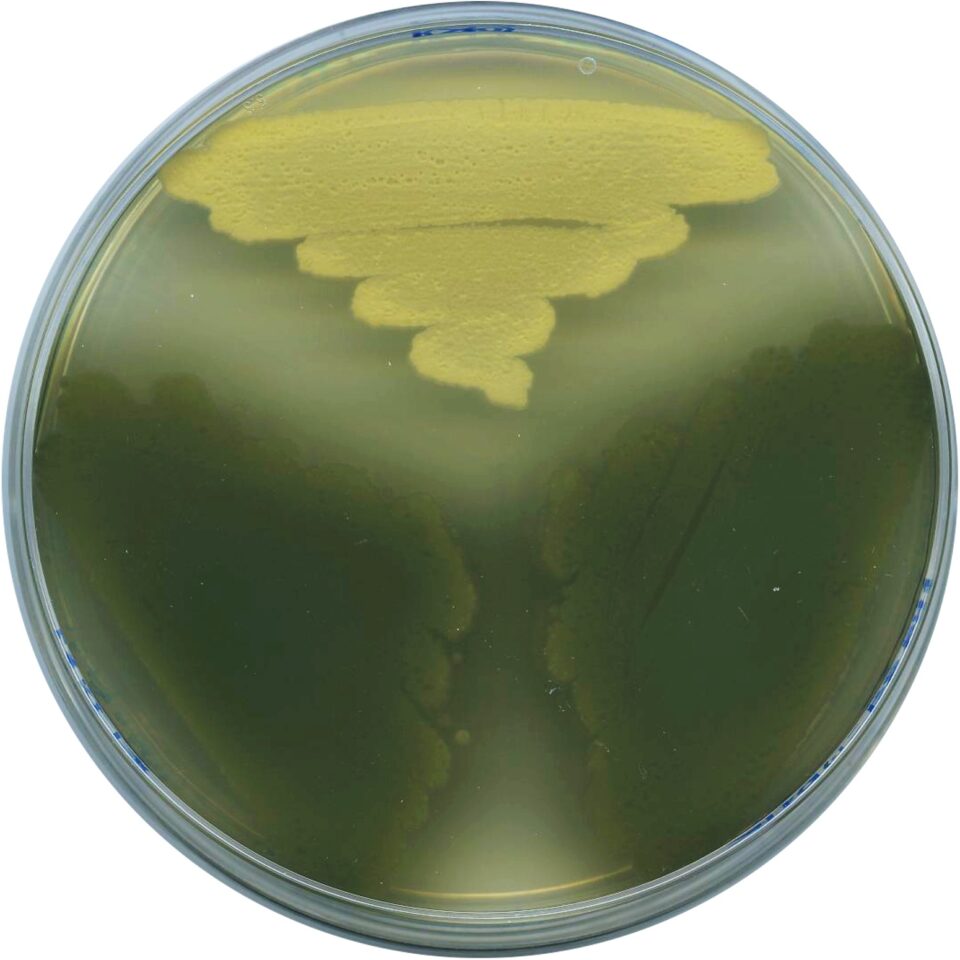
Role of Streptomyces developmental regulators in control of antibiotic production
The vast majority of natural product biosynthetic gene clusters encoded in sequenced actinobacterial genomes are cryptic, meaning that they are not produced in laboratory conditions. Crucial to unlocking this genetic potential is understanding how these bacteria regulate gene expression in response to their environments. Our lab is investigating the role of the global regulatory molecule c-di-GMP in regulation of natural product biosynthesis by diverse actinobacteria.